Main Second Level Navigation
Spotlight on Major Technology Upgrades in Donnelly Centre
Jovana Drinjakovic
In 2018, the Donnelly Centre acquired new high-end DNA sequencing instruments and microscopes and also launched the facility for mass-spectrometry analysis. While ensuring the Centre’s researchers remain at the leading edge of discovery, these advances also present new opportunities for collaboration within the Centre and other research labs in Toronto.
Donnelly Sequencing Centre gets state-of-the-art new sequencer
Founded in 2010, the Donnelly Sequencing Centre provides broad expertise in DNA and RNA sequencing technologies to labs in the Centre and external collaborators. The facility operates at cost and is the only Illumina Certified Service Provider of Next Generation Sequencing in Toronto. .
Thanks to a $5.6M grant from the Canada Foundation for Innovation and the Ontario Research, awarded to Donnelly Centre Director Brenda Andrews, the Centre was able to add a NovaSeq6000 system, a top of the line sequencer, to its suite of instruments consisting of Illumina HiSeq2500, NextSeq500, and MiSeq platforms. The new instrument can sequence 60 complete human genomes in 44 hours, at a cost of $1,500 CAD each. The DSC has also recently purchased 200 TB of additional computer server space to handle the substantial amount of increased data produced by the DSC with its new infrastructure.
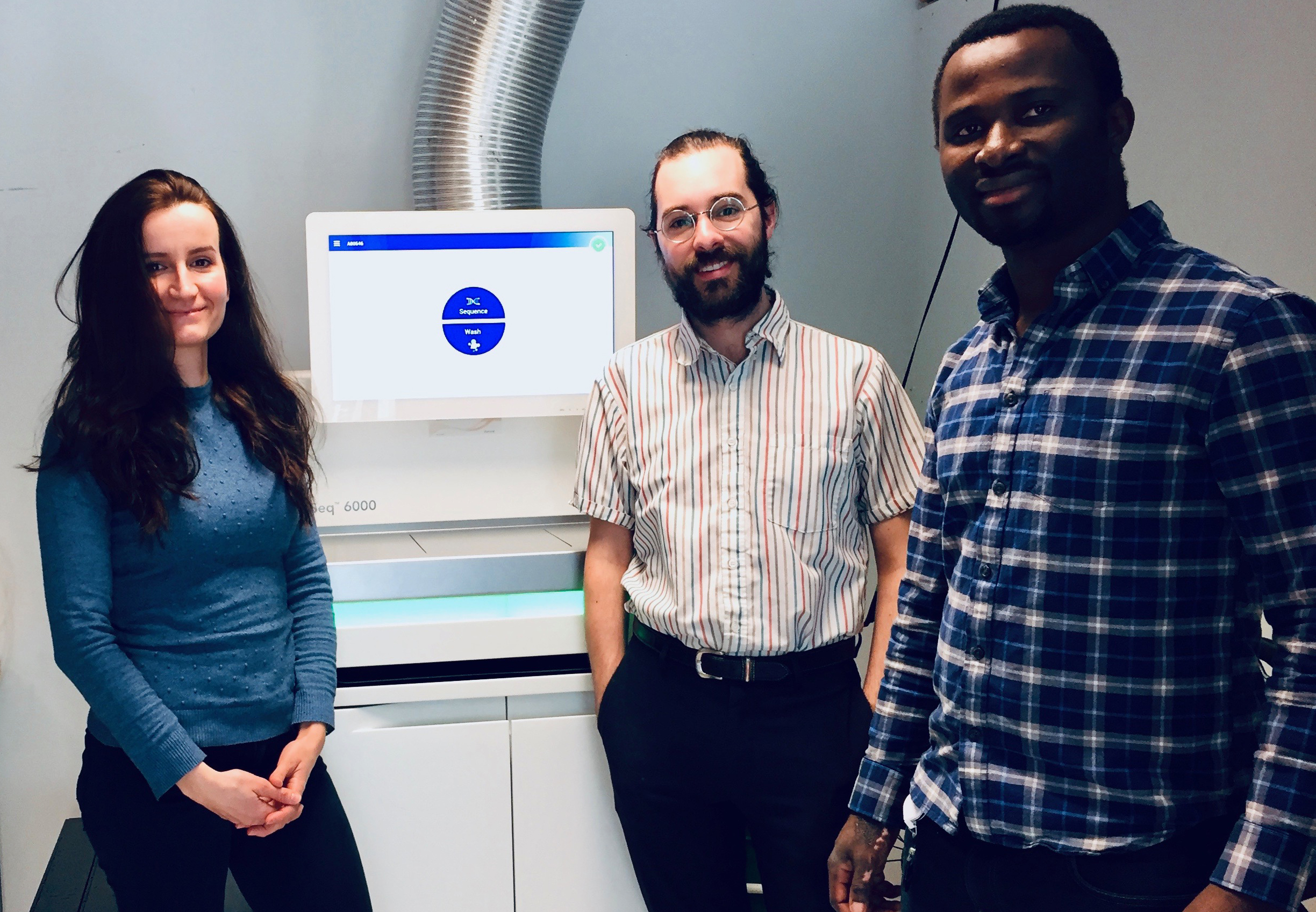
“As Director of the DSC I am very proud of the outstanding team we have assembled, including Tanja Durbic (DSC Coordinator), Kyle Turner and Michael Forbiteh, who have produced high quality sequencing data for over a hundred sequencing projects during the past year, and with record turnaround times,” says Professor Ben Blencowe, DSC Director. “A unique capability of the DSC is its ability to work with clients to develop custom sequencing protocols to foster innovative research projects. As an example, the DSC has developed custom sequencing protocols to enable new genome-wide CRISPR-based screens and this area will represent an ongoing major focus of the Centre’s work during 2019.”
In 2018 alone, the DSC helped researchers: discover a network of more than 200 genes linked to autism, through a project led by Thomas Gonatopoulos-Pournatzis in the Blencowe lab; assemble the world’s first cannabis chromosome map, led by Kaitlin Laverty in the Hughes lab; and map epigenetic changes linked to telomere maintenance and cancer, in collaboration with Uri Tabori’s lab at the Hospital for Sick Children. Overall, the DSC has ongoing projects with 22 research groups in the Centre with more than 120 external collaborations.
Other contributions, from Aaron Wheeler and Fritz Roth, of the Donnelly Centre, and Brian Cox, of U of T’s Department of Physiology, have brought in another Illumina MiSeq instrument to complement the NovaSeq6000 system, allowing the DSC team to accommodate users with smaller-scale projects, and extended the lifespan of the existing infrastructure.
“These new developments in the DSC have allowed us to simultaneously expand the scale of sequencing projects, reduce costs and turnaround times, and as a consequence maintain our competitive edge in the broader community,” says Durbic.
New imaging equipment
Donnelly investigators also leveraged the infrastructure grant described above to acquire two new live-cell imaging instruments: a fully automated Opera Phenix microscope for high throughput imaging studies and a VT-iSIM high speed super-resolution imaging system for visualizing intracellular compartments.
These new instruments, together with existing infrastructure, allow Donnelly researchers to capture thousands of images in a single day to study how genetic mutations, drugs or other environmental changes, such as nutrient availability or temperature, affect basic processes in cells: from protein trafficking, to cell aging and death. For example, the Centre’s researchers now have the ability to screen the entire genome-wide collection of yeast mutants—about 6,000 of them— in a single day under the same experimental conditions to ensure robust data collection. In addition to yeast work, ongoing studies also include work on mouse and human cells.
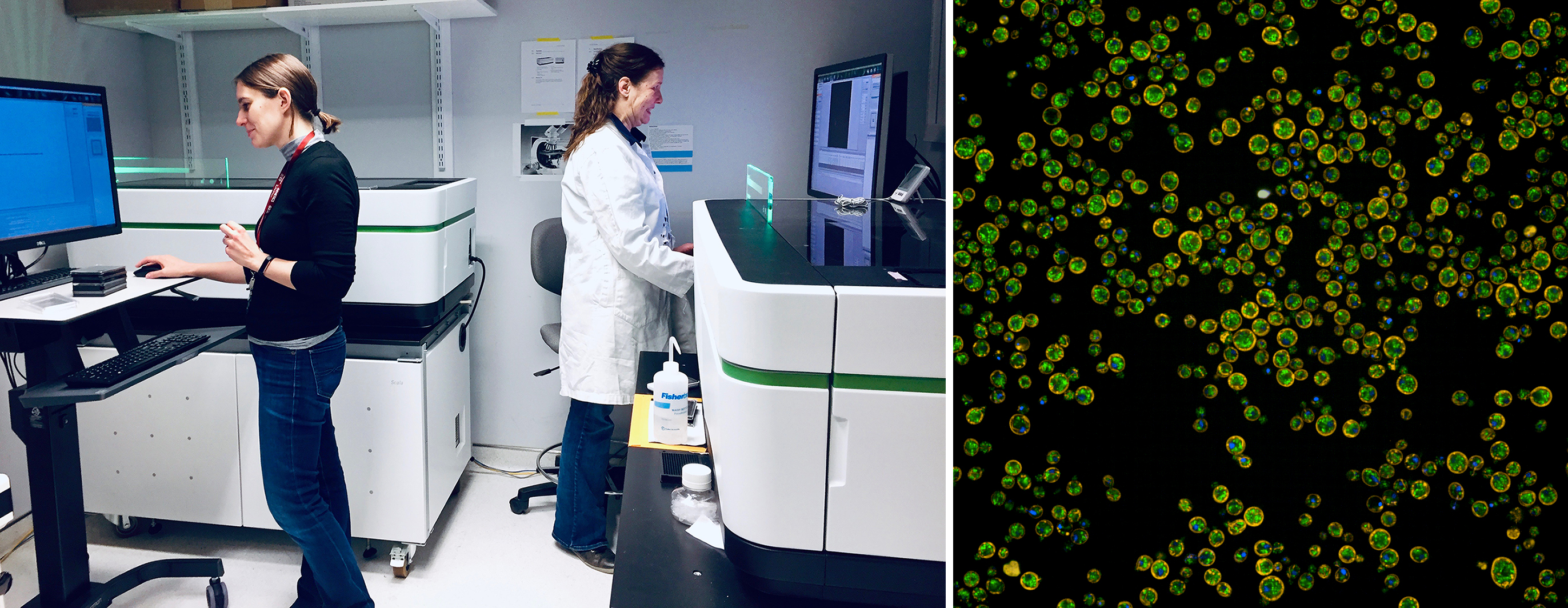
Previously, an older version of the Opera system enabled the Andrews and Boone labs to complete a landmark study that revealed where in the cell each of ~4,000 yeast proteins — almost all proteins produced by yeast— is localized and how it moves about in response to different stimuli. These large-scale image datasets have catalyzed significant collaborations with between computational and experimental biologists, to develop automated methods, involving the latest artificial intelligence algorithms, to identify important features in cell images.
The new VT-iSIM super-resolution microscope allows zooming in on subcellular structures of interest with a spatial resolution of up to 100nm, about a thousand times smaller than the width of human hair. “This new machine is also faster than standard confocal microscopes, producing images at up to 1000 frames per second, allowing us to monitor multiple fluorophores in real-time,” says Mojca Mattiazzi Usaj, a research associate in the Andrews lab who is studying how genetic and environmental changes influence morphology of the cell and more than a dozen of its inner compartments.
Newly launched Donnelly Centre facility for mass-spectrometry
Launched earlier in the year, the Donnelly Centre Mass Spec Facility offers expertise in available mass spectrometry technologies and new method development and is open to both Donnelly Centre researchers and external collaborators. The facility is directed by Donnelly Centre researchers, Jack Greenblatt and Hannes Röst, who are leading experts in mass spectrometry methods and data analysis, while Edyta Marcon oversees its daily operations.
 "Our aim is to allow different research groups to get access to high-end mass spectrometry instruments as well as expertise and knowledge that we have in the Donnelly Centre to help them answer interesting and complicated questions,” says Röst. Ongoing projects include understanding how Epstein-Barr virus infects cells, in collaboration with Lori Frappier, of U of T’s Department of Molecular Genetics, as well as detection of biomarkers for vascular disease, mapping of protein interactions in cells and detection of chemical modifications on proteins that can have profound effects on their function. Greenblatt and Röst are also working to develop methods for detecting chemical modifications on RNA molecules, which help bring DNA code to life, and which have so far remained elusive.
"Our aim is to allow different research groups to get access to high-end mass spectrometry instruments as well as expertise and knowledge that we have in the Donnelly Centre to help them answer interesting and complicated questions,” says Röst. Ongoing projects include understanding how Epstein-Barr virus infects cells, in collaboration with Lori Frappier, of U of T’s Department of Molecular Genetics, as well as detection of biomarkers for vascular disease, mapping of protein interactions in cells and detection of chemical modifications on proteins that can have profound effects on their function. Greenblatt and Röst are also working to develop methods for detecting chemical modifications on RNA molecules, which help bring DNA code to life, and which have so far remained elusive.
The facility operates at cost and has two first-rate mass spectrometry instruments which can be reconfigured for diverse research needs. With such in-house knowledge, the Centre’s researchers no longer have to seek external help making their research faster and cost-effective.
Follow us on Twitter to keep up with Donnelly Centre news.
