Main Second Level Navigation
Artem Babaian
PhD

Qualification
- University of Cambridge, Banting Postdoctoral Researcher, 2021-2022.
- Independent Researcher, 2019-2021.
- University of British Columbia, PhD in Medical Genetics, 2012-2019.
- McMaster University, BSc (Hons) in Molecular Biology and Genetics, 2007-2011.
MY RESEARCH OVERVIEW
The Laboratory for RNA-Based Lifeforms (RNAlab) at University of Toronto is a combined Computational and Molecular research team. Our passion is in understanding the interplay between RNA elements and Genetics. The primary domains where we interrogate this question is through the exploration of the vast evolution and biodiversity of Earth’s RNA viruses, and microplastic degrading enzymes.
ENTERING THE PLATINUM AGE OF VIRUS DISCOVERY (Short Lecture)
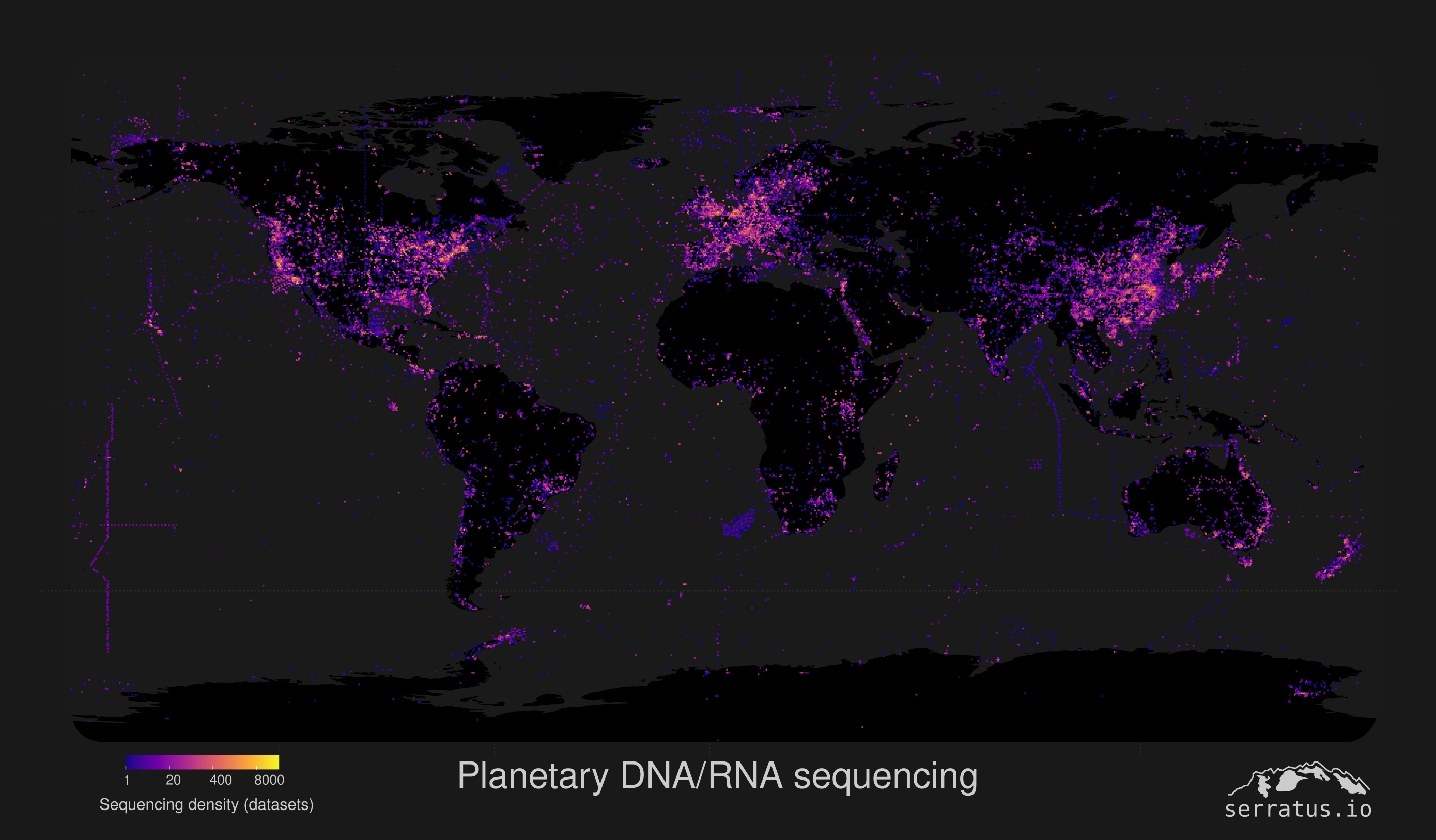
The growth of DNA and RNA sequencing data is staggering, outpacing Moore’s Law. Public databases now hold 120+ petabytes of raw sequencing data from more than 30 million samples, and this amount doubles every 24 months. Samples span from experimental cancer cells in a lab at the University of Toronto to anal swabs of wild penguins in Antarctica, and everything in between. Beyond their intended purpose, these data also hold genetic sequences from viruses and other genetic parasites in the samples. Yet they remain unanalyzed.
Our mission is to illuminate the epic tapestry of genetic diversity on Earth. To accomplish this we develop state of the art computational techniques such as Serratus and Logan which enable a new era of petabase-scale genomics. This allows us and other scientists to access and grok billions of dollars of sequence data, and investigate the evolution and ecology of genes the across our planet.
Planetary-Scale Virus Surveillance Network
For the discovery of RNA viruses: we focus on advancing the capability to find “Dark Viruses”, highly-divergent and entirely uncharacterized genetic parasites such as the Obelisks. Beyond merely discovering fringe virus and virus-like entities, we pursue understanding how these elements impact biology and health. For example, are “Dark Viruses” the cause for human diseases of unknown etiology such as Alzheimer’s Disease, Crohn’s Disease, endometriosis, or cancer?
Creating Biotechnologies for Terraforming Microplastic Contaminants
For the discovery of plastid-degrading enzymes: we focus on developing computational and molecular ultra-high throughput platforms to interrogate the evolutionary landscape of enzymes which can degrade or upcycle plastic waste. Microplastics global contaminant, we are developing the solutions which can remediate these toxins at the terraform scale.
Visit Dr. Artem Babaian's Discover Research profile to learn more.
SELECT PUBLICATIONS
-
Petabase-scale sequence alignment catalyses viral discovery. Edgar et al. Nature. 2022.
-
Hybrids of RNA viruses and viroid-like elements replicate in fungi. Forgia et al. Nature Communications. 2023.
-
Logan: Planetary-Scale Genome Assembly Surveys Life’s Diversity. Chikhi et al. Preprint. 2024.
-
A Parasite Odyssey: An RNA virus conceled in Toxoplasma gondii. Gupta et al. Virus Evolution. 2024.
-
Viroid-like colonists of human microbiomes. Zheludev et al. Cell. 2024.
