Selected Publications
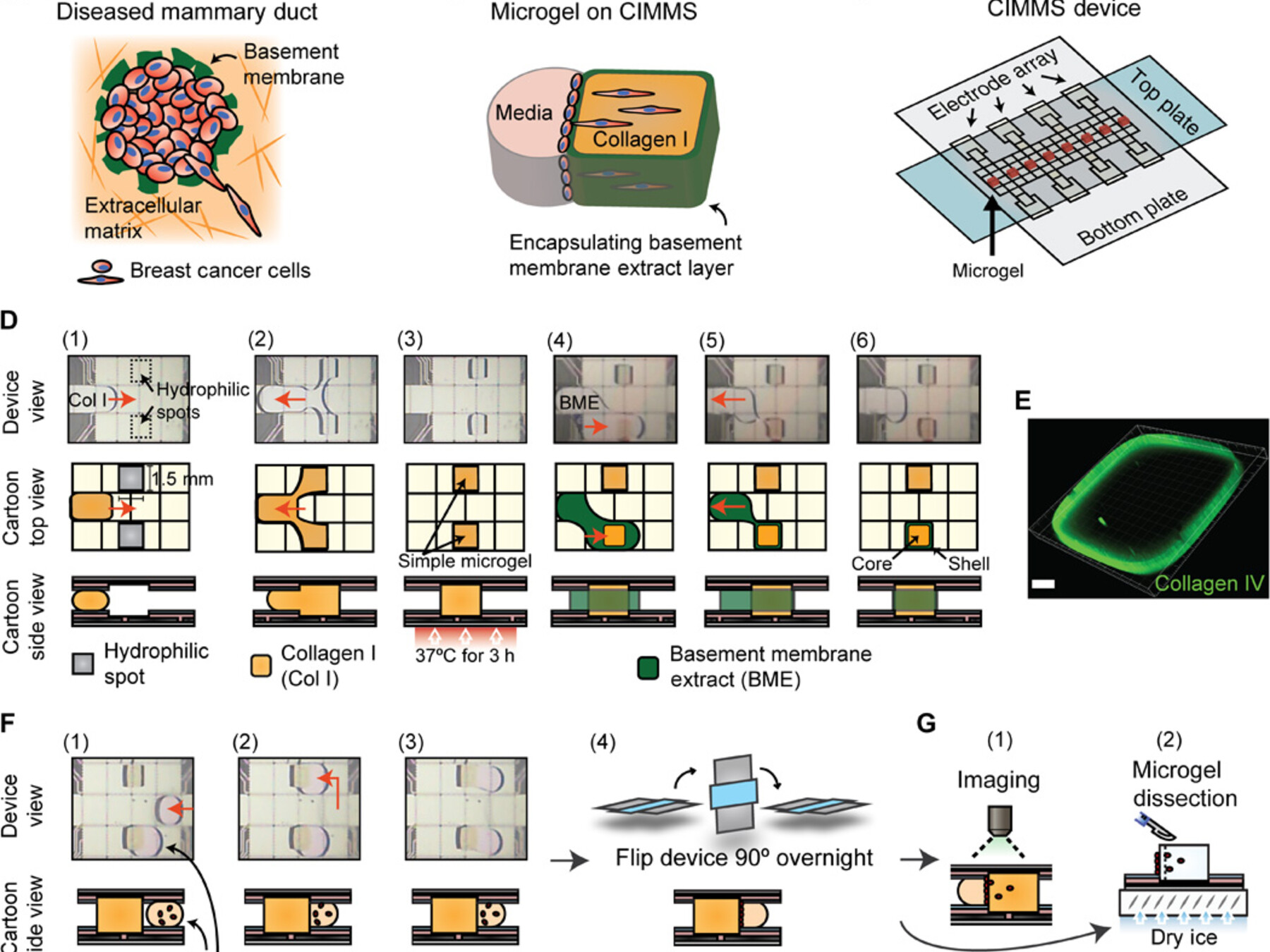
Latest feature: Cell invasion in digital microfluidic microgel systems
In this study, researchers used digital microfluidics to develop a novel in vivo-like microgel system for studying cell invasions. This system allows both imaging of extracellular matrix remodeling (an important aspect of metastasis) as well as collection of invading cell subpopulations for RNA-Seq, synthesizing the strengths of the two main existing tools for in vitro studies of cell invasion.
The study was conducted in the lab of Aaron Wheeler, based here at U of T in the Donnelly Centre. To establish the performance of this assay for yielding RNA-Seq data, and to compare expression between invading and noninvading cells, libraries were run on the NextSeq500 at the Donnelly Sequencing Centre.
Please make sure to shoot us an email when your DSC collaboration is published - we'd like to feature it!
2022
Barutcu, A. R., Wu, M., Braunschweig, U., Dyakov, B. J., Luo, Z., Turner, K. M., ... & Blencowe, B. J. (2022). Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns. Molecular Cell, 82(5), 1035-1052. [Pubmed]
Biancolin, A. D., Jeong, H., Mak, K. W., Yuan, Z., & Brubaker, P. L. (2022). Disrupted and elevated circadian secretion of glucagon-like peptide-1 in a murine model of type 2 diabetes. Endocrinology, 163(9); [Pubmed]
Erwood, S., Bily, T. M., Lequyer, J., Yan, J., Gulati, N., Brewer, R. A., ... & Cohn, R. D. (2022). Saturation variant interpretation using CRISPR prime editing. Nature Biotechnology, 1-11; [Pubmed]
Han, H., Mikolajewicz, N., Brown, K., & Moffat, J. (2022). Multi-level cellular and functional annotation of single-cell transcriptomes.[Article]
Kim, B., Sasaki, A., Murphy, K., & Matthews, S. G. (2022). DNA methylation signatures in human neonatal blood following maternal antenatal corticosteroid treatment. Translational Psychiatry, 12(1), 1-8.[Pubmed]
Kim, S. H., Steere, L., Zhang, Y. K., McGregor, C., Hahne, C., Zhou, Y., ... & Cowen, L. E. (2022). Inhibiting C-4 Methyl Sterol Oxidase with Novel Diazaborines to Target Fungal Plant Pathogens. ACS Chemical Biology, 17(6),1343–1350, [Pubmed]
Li, B. B., Scott, E. Y., Olafsen, N. E., Matthews, J., & Wheeler, A. R. (2022). Analysis of the effects of aryl hydrocarbon receptor expression on cancer cell invasion via three-dimensional microfluidic invasion assays. Lab on a Chip, 22(2), 313-325. [Pubmed]
Martchenko, A., Biancolin, A. D., Martchenko, S. E., & Brubaker, P. L. (2022). Nobiletin ameliorates high fat-induced disruptions in rhythmic glucagon-like peptide-1 secretion. Scientific reports, 12(1), 1-14.[Pubmed]
Peterson, J., McTiernan, C. D., Thome, C., Khaper, N., Lees, S. J., Boreham, D. R., ... & Tharmalingam, S. (2022). Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System. Bioengineering, 9(5), 214. [Pubmed]
Singh, S., Chokshi, C., Tieu, D., Brown, K., Venugopal, C., Rossotti, M., ... & Moffat, J. (2022). Functional discovery of targetable dependencies in recurrent glioblastoma.[Preprint]
Song, J., Nabeel-Shah, S., Pu, S., Lee, H., Braunschweig, U., Ni, Z., ... & Greenblatt, J. F. (2022). Regulation of alternative polyadenylation by the C2H2-zinc-finger protein Sp1. Molecular Cell, 82(17), 3135-3150. [Pubmed]
Khobzey, M. (2022). Exploring different approaches for constructing temperature sensitive alleles of yeast essential genes (Doctoral dissertation, University of Toronto (Canada)). [Proquest]
Ng, K. (2022). Mapping of the Novel mRNA Acetylation, ac4C, in the HEK293 Transcriptome (Doctoral dissertation, University of Toronto (Canada)). [Proquest]
2021
Kelil, A., Gallo, E., Banerjee, S., Adams, J. J., & Sidhu, S. S. (2021). CellectSeq: In silico discovery of antibodies targeting integral membrane proteins combining in situ selections and next-generation sequencing. Communications biology, 4(1), 1-13. [Pubmed]
Ma, Y., Chen, K., Xia, F., Atwal, R., Wang, H., Ahmed, S. U., ... & Kelley, S. O. (2021). Phage-Based Profiling of Rare Single Cells Using Nanoparticle-Directed Capture. ACS nano, 15(12), 19202-19210. [Pubmed]
Maino, E., Wojtal, D., Evagelou, S. L., Farheen, A., Wong, T. W., Lindsay, K., ... & Cohn, R. D. (2021). Targeted genome editing in vivo corrects a Dmd duplication restoring wild‐type dystrophin expression. EMBO molecular medicine, 13(5), e13228.[Pubmed]
Nabeel-Shah, S., Lee, H., Ahmed, N., Burke, G., Farhangmehr, S., Ashraf, K., ... & Greenblatt, J. (2021). SARS-CoV-2 nucleocapsid protein attenuates stress granules and interacts with mRNAs to impair host stress response. Iscience, 25(1):103562. [Pubmed]
Pačnik, K., Ogrizović, M., Diepold, M., Eisenberg, T., Žganjar, M., Žun, G., ... & Natter, K. (2021). Identification of novel genes involved in neutral lipid storage by quantitative trait loci analysis of Saccharomyces cerevisiae. BMC genomics, 22(1), 1-15. [Pubmed]
Stokes, K., Nunes, M., Trombley, C., Flôres, D. E., Wu, G., Taleb, Z., ... & Karpowicz, P. (2021). The circadian clock gene, BMAL1, regulates intestinal stem cell signaling and represses tumor initiation. Cellular and molecular gastroenterology and hepatology, 12(5), 1847-1872. [Pubmed]
Subramanian, A., Hall, M., Hou, H., Mufteev, M., Yu, B., Yuki, K. E., ... & Derry, W. B. (2021). Alternative polyadenylation is a determinant of oncogenic Ras function. Science advances, 7(51), eabh0562.[Pubmed]
Vaughan, C. A., Singh, S., Subler, M. A., Windle, J. J., Inoue, K., Fry, E. A., ... & Deb, S. (2021). The oncogenicity of tumor-derived mutant p53 is enhanced by the recruitment of PLK3. Nature communications, 12(1), 1-19. [Pubmed]
2020
Alerasool, N., Segal, D., Lee, H., & Taipale, M. (2020). An efficient KRAB domain for CRISPRi applications in human cells. Nature Methods, 17(11), 1093-1096.[Nature Methods]
Aregger, M., Lawson, K. A., Billmann, M., Costanzo, M., Tong, A. H., Chan, K., ... & Moffat, J. (2020). Systematic mapping of genetic interactions for de novo fatty acid synthesis identifies C12orf49 as a regulator of lipid metabolism. Nature metabolism, 2(6), 499-513.[Nature Metabolism]
Farini, D., Cesari, E., Weatheritt, R. J., La Sala, G., Naro, C., Pagliarini, V., ... & Sette, C. (2020). A dynamic splicing program ensures proper synaptic connections in the developing cerebellum. Cell Reports, 31(9), 107703. [Science Direct]
Gonatopoulos-Pournatzis, T., Niibori, R., Salter, E. W., Weatheritt, R. J., Tsang, B., Farhangmehr, S., ... & Blencowe, B. J. (2020). Autism-misregulated eIF4G microexons control synaptic translation and higher order cognitive functions. Molecular Cell, 77(6), 1176-1192. [Science Direct]
Kelil, A., Gallo, E., Adams, J., Moffat, J., & Sidhu, S. (2020). CellectSeq: In Silico Discovery of Antibodies Targeting Integral Membrane Proteins Combining In Situ Selections of Phage Displayed Synthetic Antibodies and Next-Generation Sequencing. [Preprint]
Kim, S. H., Liu, C., Zhou, Y., Zhang, Y. K., McGregor, C., Steere, L., ... & Cowen, L. E. (2020). Inhibiting protein prenylation with benzoxaboroles to target fungal plant pathogens. ACS Chemical Biology, 15(7), 1930-1941. [ACS Publications]
Lamanna, J., Scott, E. Y., Edwards, H. S., Chamberlain, M. D., Dryden, M. D., Peng, J., ... & Wheeler, A. R. (2020). Digital microfluidic isolation of single cells for-Omics. Nature communications, 11(1), 1-13. [Pubmed]
Lee, E. S., Wolf, E. J., Smith, H. W., Emili, A., & Palazzo, A. F. (2020). TPR is required for the nuclear export of mRNAs and lncRNAs from intronless and intron-poor genes. Nucleic acids research, 48(20), 11645–11663. [Pubmed]
Li, B. B., Scott, E. Y., Chamberlain, M. D., Duong, B. T., Zhang, S., Done, S. J., & Wheeler, A. R. (2020). Cell invasion in digital microfluidic microgel systems. Science advances, 6(29), eaba9589. [Link]
Martchenko, S. E., Martchenko, A., Cox, B. J., Naismith, K., Waller, A., Gurges, P., ... & Brubaker, P. L. (2020). Circadian GLP-1 secretion in mice is dependent on the intestinal microbiome for maintenance of diurnal metabolic homeostasis. Diabetes, 69(12), 2589-2602.[Pubmed]
Nabeel-Shah, S., Lee, H., Ahmed, N., Marcon, E., Farhangmehr, S., Pu, S., ... & Greenblatt, J. F. (2020). SARS-CoV-2 Nucleocapsid protein attenuates stress granule formation and alters gene expression via direct interaction with host mRNAs. BioRxiv. [Science Direct]
Oh, Y., Yang, S., Liu, X., Jana, S., Izaddoustdar, F., Gao, X., ... & Backx, P. H. (2020). Transcriptomic Bioinformatic Analyses of Atria Uncover Involvement of Pathways Related to Strain and Post-translational Modification of Collagen in Increased Atrial Fibrillation Vulnerability in Intensely Exercised Mice. Frontiers in physiology, 11, 605671. [Pubmed]
Rodrigues, D. C., Mufteev, M., Weatheritt, R. J., Djuric, U., Ha, K. C., Ross, P. J., ... & Ellis, J. (2020). Shifts in ribosome engagement impact key gene sets in neurodevelopment and ubiquitination in Rett syndrome. Cell reports, 30(12), 4179-4196. [Pubmed]
Sasaki, A., Kim, B., Murphy, K. E., & Matthews, S. G. (2020). Impact of ex vivo sample handling on DNA methylation profiles in human cord blood and neonatal dried blood spots. Frontiers in Genetics, 11, 224. [Pubmed]
Sinha, A., Saleh, A., Endersby, R., Yuan, S. H., Chokshi, C. R., Brown, K. R., ... & Katyal, S. (2020). RAD51-mediated DNA homologous recombination is independent of PTEN mutational status. Cancers, 12(11), 3178.[Pubmed]
van Leeuwen, J., Pons, C., Tan, G., Wang, J. Z., Hou, J., Weile, J., ... & Boone, C. (2020). Systematic analysis of bypass suppression of essential genes. Molecular systems biology, 16(9), e9828. [Pubmed]
Wolf, E. J., Miles, A., Lee, E. S., Nabeel-Shah, S., Greenblatt, J. F., Palazzo, A. F., ... & Emili, A. (2020). MKRN2 physically interacts with GLE1 to regulate mRNA export and zebrafish retinal development. Cell Reports, 31(8), 107693.[Pubmed]
Reid CG. Investigating the molecular basis for cooperative behaviour in a mutualistic plant-ant. Master's Thesis, University of Toronto. 2020; [TSpace]
Zhai AH. Structural and Functional Characterization of the RING-like Protein RTF2. Doctoral Thesis, University of Toronto. 2020; [TSpace]
2019
Garg, J., Saettone, A., Nabeel-Shah, S., Cadorin, M., Ponce, M., Marquez, S., ...& Fillingham J. (2019). The Med31 conserved component of the divergent mediator complex in Tetrahymena thermophila participates in developmental regulation. Current Biology, 29(14), 2371–2379.e6. [Pubmed]
Laverty, K. U., Stout, J. M., Sullivan, M. J., Shah, H., Gill, N., Holbrook, L., ... & van Bakel, H. (2019). A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome research, 29(1), 146–156.. [Pubmed]
Magomedova, L., Tiefenbach, J., Zilberman, E., Le Billan, F., Voisin, V., Saikali, M., …& Cummins, C. L. (2019). ARGLU1 is a transcriptional coactivator and splicing regulator important for stress hormone signaling and development. Nucleic acids research, 47(6), 2856–2870.[Pubmed]
Mair, B., Tomic, J., Masud, S. N., Tonge, P., Weiss, A., Usaj, M., ...& Moffat, J. (2019). Essential Gene Profiles for Human Pluripotent Stem Cells Identify Uncharacterized Genes and Substrate Dependencies. Cell reports, 27(2), 599–615.e12. [Pubmed]
McAuley, S., Huynh, A., Howells, A., Walpole, C., Maxwell, A., & Nodwell, J. R. (2019). Discovery of a novel DNA gyrase-targeting antibiotic through the chemical perturbation of Streptomyces venezuelae sporulation. Cell chemical biology, 26(9), 1274-1282[Pubmed]
Saettone, A., Ponce, M., Nabeel-Shah, S., Fillingham, J.(2019). RACS: rapid analysis of ChIP-Seq data for contig based genomes. BMC Bioinformatics. 2019; 20(533); [Pubmed]
Sin-Chan, P., Mumal, I., Suwal, T., Ho, B., Fan, X., Singh, I., …& Huang, A. (2019). A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor. Cancer cell, 36(1), 51–67.e7 [Pubmed]
Vinals, D. F., Kitov, P. I., Tu, Z., Zou, C., Cairo, C. W., Lin, H. C. H., & Derda, R. (2019). Selection of galectin‐3 ligands derived from genetically encoded glycopeptide libraries. Peptide Science, 111(1), e24097. [Pubmed]
Xiao, H., Li, C., Al Rwahnih, M., Dolja, V., & Meng, B. (2019). Metagenomic analysis of riesling grapevine reveals a complex virome including two new and divergent variants of grapevine leafroll-associated virus 3. Plant disease, 103(6), 1275-1285. [Pubmed]
Zhao, Y., Rafatian, N., Feric, N. T., Cox, B. J., Aschar-Sobbi, R., Wang, E. Y., ... & Radisic, M. (2019). A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell, 176(4), 913-927. [Pubmed]
Ng, M. K. (2019). Characterization of Nucleosomes Containing Specific Forms of the Histone Variant H2A. Z. Doctoral Thesis, York University.[Yorkspace]
2018
Davis GM, Tu S, Anderson JW, Colson RN, Gunzburg MJ, Francisco MA, Ray D, Shrubsole SP, Sobotka JA, Seroussi U, Lao RX, Maity T, Wu MZ, McJunkin K, Morris QD, Hughes TR, Wilce JA, Claycomb J, Weng Z, Boag PR. The TRIM-NHL protein NHL-2 is a co-factor in the nuclear and somatic RNAi pathways in C. elegans. e. eLife. 2018; 7:. [Pubmed]
Ma YV, Xu L, Mei X, Middleton K, You L. Mechanically stimulated osteocytes reduce the bone-metastatic potential of breast cancer cells in vitro by signaling through endothelial cells. Journal of Cellular Biochemistry. 2018; [Pubmed]
Gonatopoulos-Pournatzis T, Wu M, Braunschweig U, Roth J, Han H, Best AJ, Raj B, Aregger M, O'Hanlon D, Ellis JD, Calarco JA, Moffat J, Gingras AC, Blencowe BJ. Genome-wide CRISPR-Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Molecular Cell. 2018; 72(3):510-524.e12. [Pubmed]
Ng S, Bennett NJ, Schulze J, Gao N, Rademacher C, Derda R. Genetically-encoded fragment-based discovery of glycopeptide ligands for DC-SIGN. Bioorganic & Medicinal Chemistry. 2018; 26(19):5368-5377. [Pubmed]
Sterne-Weiler T, Weatheritt RJ, Best AJ, Ha KCH, Blencowe BJ. Efficient and accurate quantitative profiling of alternative splicing patterns of any complexity on a laptop. Molecular Cell. 2018; 72(1):187-200.e6. [Pubmed]
Veri AO, Miao Z, Shapiro RS, Tebbji F, O'Meara TR, Kim SH, Colazo J, Tan K, Vyas VK, Whiteway M, Robbins N, Wong KH, Cowen LE. Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS genetics. 2018; 14(3):e1007270. [Pubmed]
Midhat U, Ting MKY, Teresinski HJ, Snedden WA. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Molecular Biology. 2018; 96(4-5):375-392. [Pubmed]
2017
Cai Q, Ye X, Chen B, Zhang B. Complete genome sequence of Exiguobacterium sp. strain N4-1P, a psychrophilic bioemulsifier producer isolated from a cold marine environment in North Atlantic Canada. Genome Announcements. 2017; 5(44):. [Pubmed]
Ilves KL, Torti D, López-Fernández H. Exon-based phylogenomics strengthens the phylogeny of Neotropical cichlids and identifies remaining conflicting clades (Cichliformes: Cichlidae: Cichlinae). Molecular Phylogenetics and Evolution. 2018; 118:232-243. [Pubmed]
Moisiadis VG, Constantinof A, Kostaki A, Szyf M, Matthews SG. Prenatal glucocorticoid exposure modifies endocrine function and behaviour for 3 generations following maternal and paternal transmission. Scientific Reports. 2017; 7(1):11814. [Pubmed]
Loll-Krippleber R, Brown GW. P-body proteins regulate transcriptional rewiring to promote DNA replication stress resistance. Nature Communications. 2017; 8(1):558. [Pubmed]
Xiong X, Bales ES, Ir D, Robertson CE, McManaman JL, Frank DN, Parkinson J. Perilipin-2 modulates dietary fat-induced microbial global gene expression profiles in the mouse intestine. Microbiome. 2017; 5(1):117. [Pubmed]
Tapial J, Ha KCH, Sterne-Weiler T, Gohr A, Braunschweig U, Hermoso-Pulido A, Quesnel-Vallières M, Permanyer J, Sodaei R, Marquez Y, Cozzuto L, Wang X, Gómez-Velázquez M, Rayon T, Manzanares M, Ponomarenko J, Blencowe BJ, Irimia M. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Research. 2017; 27(10):1759-1768. [Pubmed]
Gueroussov S, Weatheritt RJ, O'Hanlon D, Lin ZY, Narula A, Gingras AC, Blencowe BJ. Regulatory expansion in mammals of multivalent hnRNP assemblies that globally control alternative splicing. Cell. 2017; 170(2):324-339.e23. [Pubmed]
Loiselle JJ, Roy JG, Sutherland LC. RBM10 promotes transformation-associated processes in small cell lung cancer and is directly regulated by RBM5. PLoS One. 2017; 12(6):e0180258. [Pubmed]
Akkouche A, Mugat B, Barckmann B, Varela-Chavez C, Li B, Raffel R, Pélisson A, Chambeyron S. Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Molecular Cell. 2017; 66(3):411-419.e4. [Pubmed]
Vaughan CA, Singh S, Grossman SR, Windle B, Deb SP, Deb S. Gain-of-function p53 activates multiple signaling pathways to induce oncogenicity in lung cancer cells. Molecular Oncology. 2017; 11(6):696-711. [Pubmed]
Singh S, Vaughan CA, Frum RA, Grossman SR, Deb S, Palit Deb S. Mutant p53 establishes targetable tumor dependency by promoting unscheduled replication. The Journal of Clinical Investigation. 2017; 127(5):1839-1855. [Pubmed]
AlZahal O, Li F, Guan LL, Walker ND, McBride BW. Factors influencing ruminal bacterial community diversity and composition and microbial fibrolytic enzyme abundance in lactating dairy cows with a focus on the role of active dry yeast. Journal of Dairy Science. 2017; 100(6):4377-4393. [Pubmed]
Burns AR, Bagg R, Yeo M, Luciani GM, Schertzberg M, Fraser AG, Roy PJ. The novel nematicide wact-86 interacts with aldicarb to kill nematodes. PLoS Neglected Tropical Diseases. 2017; 11(4):e0005502. [Pubmed]
Han H, Braunschweig U, Gonatopoulos-Pournatzis T, Weatheritt RJ, Hirsch CL, Ha KCH, Radovani E, Nabeel-Shah S, Sterne-Weiler T, Wang J, O'Hanlon D, Pan Q, Ray D, Zheng H, Vizeacoumar F, Datti A, Magomedova L, Cummins CL, Hughes TR, Greenblatt JF, Wrana JL, Moffat J, Blencowe BJ. Multilayered control of alternative splicing regulatory networks by transcription factors. Molecular Cell. 2017; 65(3):539-553.e7. [Pubmed]
Mentasti M, Cassier P, David S, Ginevra C, Gomez-Valero L, Underwood A, Afshar B, Etienne J, Parkhill J, Chalker V, Buchrieser C, Harrison TG, Jarraud S; ESCMID Study Group for Legionella Infections (ESGLI). Rapid detection and evolutionary analysis of Legionella pneumophila serogroup 1 sequence type 47. Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. 2017; 23(4):264.e1-264.e9. [Pubmed]
Leach MD, Kim T, DiGregorio SE, Collins C, Zhang Z, Duennwald ML, Cowen LE. Candida albicans is resistant to polyglutamine aggregation and toxicity. G3 (Bethesda, MD.). 2017; 7(1):95-108. [Pubmed]
2016
Quesnel-Vallières M, Dargaei Z, Irimia M, Gonatopoulos-Pournatzis T, Ip JY, Wu M, Sterne-Weiler T, Nakagawa S, Woodin MA, Blencowe BJ, Cordes SP. Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Molecular Cell. 2016; 64(6):1023-1034. [Pubmed]
Soloveychik M, Xu M, Zaslaver O, Lee K, Narula A, Jiang R, Rosebrock AP, Caudy AA, Meneghini MD. Mitochondrial control through nutritionally regulated global histone H3 lysine-4 demethylation. Scientific Reports. 2016; 6:37942. [Pubmed]
Lee JA, Robbins N, Xie JL, Ketela T, Cowen LE. Functional genomic analysis of Candida albicans adherence reveals a key role for the Arp2/3 complex in cell wall remodelling and biofilm formation. Molecular Cell. 2016; 12(11):e1006452. [Pubmed]
Schmitges FW, Radovani E, Najafabadi HS, Barazandeh M, Campitelli LF, Yin Y, Jolma A, Zhong G, Guo H, Kanagalingam T, Dai WF, Taipale J, Emili A, Greenblatt JF, Hughes TR. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Research. 2016; 26(12):1742-1752. [Pubmed]
van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Ušaj MM, Pechlaner M, Takar M, Ušaj M1 VanderSluis B, Andrusiak K, Bansal P, Baryshnikova A, Boone CE, Cao J, Cote A, Gebbia M, Horecka G, Horecka I, Kuzmin E, Legro N, Liang W, van Lieshout N, McNee M, San Luis BJ, Shaeri F, Shuteriqi E, Sun S, Yang L, Youn JY, Yuen M, Costanzo M, Gingras AC, Aloy P, Oostenbrink C, Murray A, Graham TR, Myers CL, Andrews BJ, Roth FP, Boone C. Exploring genetic suppression interactions on a global scale. Science (New York, N.Y.). 2016; 354(6312). [Pubmed]
Liang L, Gnaneshan S, Garduño RA, Mallo GV. Genome sequence of Listeria monocytogenes Plasmid pLM-C-273 carrying genes related to stress resistance. Genome Announcements. 2016; 4(5):. [Pubmed]
Shekhar-Guturja T, Gunaherath GM, Wijeratne EM, Lambert JP, Averette AF, Lee SC, Kim T, Bahn YS, Tripodi F, Ammar R, Döhl K, Niewola-Staszkowska K, Schmitt L, Loewith RJ, Roth FP, Sanglard D, Andes D, Nislow C, Coccetti P, Gingras AC, Heitman J, Gunatilaka AA, Cowen LE. Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nature Chemical Biology. 2016; 12(10):867-75. [Pubmed]
Tsalikis J, Pan Q, Tattoli I, Maisonneuve C, Blencowe BJ, Philpott DJ, Girardin SE. The transcriptional and splicing landscape of intestinal organoids undergoing nutrient starvation or endoplasmic reticulum stress. BMC Genomics. 2016; 17:680. [Pubmed]
Teatero S, Lemire P, Dewar K, Wasserscheid J, Calzas C, Mallo GV, Li A, Athey TB, Segura M, Fittipaldi N. Genomic recombination leading to decreased virulence of group B Streptococcus in a mouse model of adult invasive disease. Pathogens (Basel, Switzerland). 2016; 5(3):. [Pubmed]
Wang Y, White MM, Moncalvo JM. Draft genome sequence of Capniomyces stellatus, the obligate gut fungal symbiont of stonefly. Genome Announcements. 2016; 4(4):. [Pubmed]
Rodor J, Pan Q, Blencowe BJ, Eyras E, Cáceres JF. The RNA-binding profile of Acinus, a peripheral component of the exon junction complex, reveals its role in splicing regulation. RNA (New York, N.Y.). 2016; 22(9):1411-26. [Pubmed]
Ip JY, Sone M, Nashiki C, Pan Q, Kitaichi K, Yanaka K, Abe T, Takao K, Miyakawa T, Blencowe BJ, Nakagawa S. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Scientific Reports. 2016; 6:27204. [Pubmed]
Stubenhaus BM, Dustin JP, Neverett ER, Beaudry MS, Nadeau LE, Burk-McCoy E, He X, Pearson BJ, Pellettieri J. Light-induced depigmentation in planarians models the pathophysiology of acute porphyrias. eLife. 2016; 5:. [Pubmed]
Wisnovsky S, Jean SR, Kelley SO. Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nature Chemical Biology. 2016; 12(7):567-73. [Pubmed]
Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. Global mapping of human RNA-RNA interactions. Molecular Cell. 2016; 62(4):618-26. [Pubmed]
Labrie V, Buske OJ, Oh E, Jeremian R, Ptak C, Gasiūnas G, Maleckas A, Petereit R, Žvirbliene A, Adamonis K, Kriukienė E, Koncevičius K, Gordevičius J, Nair A, Zhang A, Ebrahimi S, Oh G, Šikšnys V, Kupčinskas L, Brudno M, Petronis A. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nature Structural & Molecular Biology. 2016; 23(6):566-73. [Pubmed]
Herman AC, Schoen DJ. Recent selection for self-compatibility in a population of Leavenworthia alabamica. Evolution; International Journal of Organic Evolution. 2016; 70(6):1212-24. [Pubmed]
D'Souza SA, Rajendran L, Bagg R, Barbier L, van Pel DM, Moshiri H, Roy PJ. The MADD-3 LAMMER kinase interacts with a p38 MAP kinase pathway to regulate the display of the EVA-1 guidance receptor in Caenorhabditis elegans. Molecular Cell. 2016; 12(4):e1006010. [Pubmed]
Yachie N, Petsalaki E, Mellor JC, Weile J, Jacob Y, Verby M, Ozturk SB, Li S, Cote AG, Mosca R, Knapp JJ, Ko M, Yu A, Gebbia M, Sahni N, Yi S, Tyagi T, Sheykhkarimli D, Roth JF, Wong C, Musa L, Snider J, Liu YC, Yu H, Braun P, Stagljar I, Hao T, Calderwood MA, Pelletier L, Aloy P, Hill DE, Vidal M, Roth FP. Pooled-matrix protein interaction screens using Barcode Fusion Genetics. Molecular Systems Biology. 2016; 12(4):863. [Pubmed]
Winick-Ng W, Caetano FA, Winick-Ng J, Morey TM, Heit B, Rylett RJ. 82-kDa choline acetyltransferase and SATB1 localize to β-amyloid induced matrix attachment regions. Scientific Reports. 2016; 6:23914. [Pubmed]
Currie KW, Brown DD, Zhu S, Xu C, Voisin V, Bader GD, Pearson BJ. HOX gene complement and expression in the planarian Schmidtea mediterranea. EvoDevo. 2016; 7:7. [Pubmed]
Rao C, Guyard C, Pelaz C, Wasserscheid J, Bondy-Denomy J1 Dewar K, Ensminger AW. Active and adaptive Legionella CRISPR-Cas reveals a recurrent challenge to the pathogen. Cellular Microbiology. 2016; 18(10):1319-38. [Pubmed]
Gnaneshan S, Hsueh YC, Liang L, Teatero S, Fittipaldi N, Mallo GV. Genome sequence of Listeria monocytogenes strain F6540 (sequence type 360) collected from food samples in Ontario, Canada. Genome Announcements. 2016; 4(1). [Pubmed]
Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, Vedadi M, Min J, Pawson TJ, Blencowe BJ, Greenblatt JF. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016; 529(7584):48-53. [Pubmed]
Tjhung KF, Kitov PI, Ng S, Kitova EN, Deng L, Klassen JS, Derda R. Silent encoding of chemical post-translational modifications in phage-displayed libraries. Journal of the American Chemical Society. 2016; 138(1):32-5. [Pubmed]
Dibitetto D, Ferrari M, Rawal CC, Balint A, Kim T, Zhang Z, Smolka MB, Brown GW, Marini F, Pellicioli A. Slx4 and Rtt107 control checkpoint signalling and DNA resection at double-strand breaks. Nucleic Acids Research. 2016; 44(2):669-82. [Pubmed]
Gonorazky H, Liang M, Cummings B, Lek M, Micallef J, Hawkins C, Basran R, Cohn R, Wilson MD, MacArthur D, Marshall CR, Ray PN, Dowling JJ. RNAseq analysis for the diagnosis of muscular dystrophy. Annals of Clinical and Translational Neurology. 2016; 3(1):55-60. [Pubmed]
2015
Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015; 163(6):1515-26. [Pubmed]
Giampietro C, Deflorian G, Gallo S, Di Matteo A, Pradella D, Bonomi S, Belloni E, Nyqvist D, Quaranta V, Confalonieri S, Bertalot G, Orsenigo F, Pisati F, Ferrero E, Biamonti G, Fredrickx E, Taveggia C, Wyatt CD, Irimia M, Di Fiore PP, Blencowe BJ, Dejana E, Ghigna C. The alternative splicing factor Nova2 regulates vascular development and lumen formation. Nature Communications. 2015; 6:8479. [Pubmed]
Jablonowski CM, Cussiol JR, Oberly S, Yimit A, Balint A, Kim T, Zhang Z, Brown GW, Smolka MB. Termination of replication stress signaling via concerted action of the Slx4 scaffold and the PP4 phosphatase. Genetics. 2015; 201(3):937-49. [Pubmed]
van Leeuwen J, Andrews B, Boone C, Tan G. Construction of Multifragment Plasmids by Homologous Recombination in Yeast. Cold Spring Harbor Protocols. 2015; 2015(9):pdb.top084111. [Pubmed]
van Leeuwen J, Andrews B, Boone C, Tan G. Rapid and Efficient Plasmid Construction by Homologous Recombination in Yeast. Cold Spring Harbor Protocols. 2015; 2015(9):pdb.prot085100. [Pubmed]
Gueroussov S, Gonatopoulos-Pournatzis T, Irimia M, Raj B, Lin ZY, Gingras AC, Blencowe BJ. An alternative splicing event amplifies evolutionary differences between vertebrates. Science (New York, N.Y.). 2015; 349(6250):868-73. [Pubmed]
Tomsic J, He H, Akagi K, Liyanarachchi S, Pan Q, Bertani B, Nagy R, Symer DE, Blencowe BJ, de la Chapelle A. A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition. Scientific Reports. 2015; 5:10566. [Pubmed]
Bagherie-Lachidan M, Reginensi A, Pan Q, Zaveri HP, Scott DA, Blencowe BJ, Helmbacher F, McNeill H. Stromal Fat4 acts non-autonomously with Dchs1/2 to restrict the nephron progenitor pool. Development (Cambridge, England). 2015; 142(15):2564-73. [Pubmed]
Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ. A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife. 2015; 4. [Pubmed]
Balint A, Kim T, Gallo D, Cussiol JR, Bastos de Oliveira FM, Yimit A, Ou J, Nakato R, Gurevich A, Shirahige K, Smolka MB, Zhang Z, Brown GW. Assembly of Slx4 signaling complexes behind DNA replication forks. The EMBO Journal. 2015; 34(16):2182-97. [Pubmed]
Burns AR, Luciani GM, Musso G, Bagg R, Yeo M, Zhang Y, Rajendran L, Glavin J, Hunter R, Redman E, Stasiuk S, Schertzberg M, Angus McQuibban G, Caffrey CR, Cutler SR, Tyers M, Giaever G, Nislow C, Fraser AG, MacRae CA, Gilleard J, Roy PJ. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nature Communications. 2015; 6:7485. [Pubmed]
Pal M, Ebrahimi S, Oh G, Khare T, Zhang A, Kaminsky ZA, Wang SC, Petronis A. High precision DNA modification analysis of HCG9 in major psychosis. Schizophrenia Bulletin. 2016; 42(1):170-7. [Pubmed]
Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Research. 2015; 4:17. [Pubmed]
Cussiol JR, Jablonowski CM, Yimit A, Brown GW, Smolka MB. Dampening DNA damage checkpoint signalling via coordinated BRCT domain interactions. The EMBO Journal. 2015; 34(12):1704-17. [Pubmed]
Quesnel-Vallières M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes & Development. 2015; 29(7):746-59. [Pubmed]
O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nature Communications. 2015; 6:6741. [Pubmed]
Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, Greenblatt J, Frey BJ, Hughes TR. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nature Biotechnology. 2015; 33(5):555-62. [Pubmed]
2014
Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014; 159(7):1511-23. [Pubmed]
Ali SS, Soo J, Rao C, Leung AS, Ngai DH, Ensminger AW, Navarre WW. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathogens. 2014; 10(11):e1004500. [Pubmed]
Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O'Hanlon D, Lin ZY, Chen GI, Easton LE, Ule J, Gingras AC, Eyras E, Blencowe BJ. A global regulatory mechanism for activating an exon network required for neurogenesis. Molecular Cell. 2014; 56(1):90-103. [Pubmed]
Mansouri S, Pan Q, Blencowe BJ, Claycomb JM, Frappier L. Epstein-Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. Journal of Virology. 2014; 88(19):11166-77. [Pubmed]
Huang SS, Banner D, Paquette SG, Leon AJ, Kelvin AA, Kelvin DJ. Pathogenic influenza B virus in the ferret model establishes lower respiratory tract infection. The Journal of General Virology. 2014; 95(Pt 10):2127-39. [Pubmed]
Marcon E, Ni Z, Pu S, Turinsky AL, Trimble SS, Olsen JB, Silverman-Gavrila R, Silverman-Gavrila L, Phanse S, Guo H, Zhong G, Guo X, Young P, Bailey S, Roudeva D, Zhao D, Hewel J, Li J, Gräslund S, Paduch M, Kossiakoff AA, Lupien M, Emili A, Wodak SJ, Greenblatt J. Human-chromatin-related protein interactions identify a demethylase complex required for chromosome segregation. Cell Reports. 2014; 8(1):297-310. [Pubmed]
Arnoldo A, Kittanakom S, Heisler LE, Mak AB, Shukalyuk AI, Torti D, Moffat J, Giaever G, Nislow C. A genome scale overexpression screen to reveal drug activity in human cells. Genome Medicine. 2014; 6(4):32. [Pubmed]
Parts L, Liu YC, Tekkedil MM, Steinmetz LM, Caudy AA, Fraser AG, Boone C, Andrews BJ, Rosebrock AP. Heritability and genetic basis of protein level variation in an outbred population. Genome Research. 2014; 24(8):1363-70. [Pubmed]
Larriba E, Jaime MD, Carbonell-Caballero J, Conesa A, Dopazo J, Nislow C, Martín-Nieto J, Lopez-Llorca LV. Sequencing and functional analysis of the genome of a nematode egg-parasitic fungus, Pochonia chlamydosporia. Fungal Genetics and Biology: FG&B. 2014; 65:69-80. [Pubmed]
Lin AY, Pearson BJ. Planarian yorkie/YAP functions to integrate adult stem cell proliferation, organ homeostasis and maintenance of axial patterning. Development (Cambridge, England). 2014; 141(6):1197-208. [Pubmed]
Ilves KL, López-Fernández H. A targeted next-generation sequencing toolkit for exon-based cichlid phylogenomics. Molecular Ecology Resources. 2014; 14(4):802-11. [Pubmed]
Matochko WL, Cory Li S, Tang SK, Derda R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Research. 2014; 42(3):1784-98. [Pubmed]
Kurat CF, Recht J, Radovani E, Durbic T, Andrews B, Fillingham J. Regulation of histone gene transcription in yeast. Cellular and Molecular Life Sciences: CMLS. 2014; 71(4):599-613. [Pubmed]
2013
Kittanakom S, Arnoldo A, Brown KR, Wallace I, Kunavisarut T, Torti D, Heisler LE, Surendra A, Moffat J, Giaever G, Nislow C. Miniature short hairpin RNA screens to characterize antiproliferative drugs. G3 (Bethesda, Md.). 2013; 3(8):1375-87. [Pubmed]
Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, Wang E, Trcka D, Thompson T, O'Hanlon D, Slobodeniuc V, Barbosa-Morais NL, Burge CB, Moffat J, Frey BJ, Nagy A, Ellis J, Wrana JL, Blencowe BJ. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013; 498(7453):241-5. [Pubmed]
Rao C, Benhabib H, Ensminger AW. Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS One. 2013; 8(5):e64129. [Pubmed]
Hill JA, Ammar R, Torti D, Nislow C, Cowen LE. Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations. Molecular Cell. 2013; 9(4):e1003390. [Pubmed]
Li Y, Zhao DY, Greenblatt JF, Zhang Z. RIPSeeker: A statistical package for identifying protein-associated transcripts from RIP-seq experiments. Nucleic Acids Research. 2013; 41(8):e94. [Pubmed]
Tompsett AR, Wiseman S, Higley E, Giesy JP, Hecker M. Effects of exposure to 17α-ethynylestradiol during larval development on growth, sexual differentiation, and abundances of transcripts in the liver of the wood frog (Lithobates sylvaticus). Aquatic Toxicology (Amsterdam, Netherlands). 2013; 126:42-51. [Pubmed]
Zhu SJ, Pearson BJ. The Retinoblastoma pathway regulates stem cell proliferation in freshwater planarians. Developmental Biology. 2013; 373(2):442-52. [Pubmed]
2012
Ammar R, Torti D, Tsui K, Gebbia M, Durbic T, Bader GD, Giaever G, Nislow C. Chromatin is an ancient innovation conserved between Archaea and Eukarya. eLife. 2012; 1:e00078. [Pubmed]
Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, Lee AY, Ryan O, Göttert H, Surendra A, van Dyk D, Giaever G, Boone C, Nislow C, Andrews BJ. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda, Md.). 2012; 2(10):1279-89. [Pubmed]
Smith AM, Durbic T, Kittanakom S, Giaever G, Nislow C. Barcode sequencing for understanding drug-gene interactions. Methods in Molecular Biology (Clifton, N.J.). 2012; 910:55-69. [Pubmed]
Onodera NT, Ryu J, Durbic T, Nislow C, Archibald JM, Rohde JR. Genome sequence of Shigella flexneri serotype 5a strain M90T Sm. Journal of Bacteriology. 2012; 194(11):3022. [Pubmed]
Kim T, Tyndel MS, Huang H, Sidhu SS, Bader GD, Gfeller D, Kim PM. MUSI: An integrated system for identifying multiple specificity from very large peptide or nucleic acid data sets. Nucleic Acids Research. 2012; 40(6):e47. [Pubmed]
Tsui K, Durbic T, Gebbia M, Nislow C. Genomic approaches for determining nucleosome occupancy in yeast. Methods in Molecular Biology (Clifton, N.J.). 2012; 833:389-411. [Pubmed]
2011
van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE. The draft genome and transcriptome of Cannabis sativa. Genome Biology. 2011; 12(10):R102. [Pubmed]
Han H, Nutiu R, Moffat J, Blencowe BJ. SnapShot: High-throughput sequencing applications. Cell. 2011; 146(6):1044, 1044.e1-2. [Pubmed]
Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011; 147(1):132-46. [Pubmed]
Smith AM, Durbic T, Oh J, Urbanus M, Proctor M, Heisler LE, Giaever G, Nislow C. Competitive genomic screens of barcoded yeast libraries. Journal of Visualized Experiments: JoVE. 2011; [Pubmed]
Ketela T, Heisler LE, Brown KR, Ammar R, Kasimer D, Surendra A, Ericson E, Blakely K, Karamboulas D, Smith AM, Durbic T, Arnoldo A, Cheung-Ong K, Koh JL, Gopal S, Cowley GS, Yang X, Grenier JK, Giaever G, Root DE, Moffat J, Nislow C. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics. 2011; 12:213. [Pubmed]
