Main Second Level Navigation
Grant Brown
PhD

Qualification
- Johns Hopkins University School of Medicine, Baltimore, MD, U.S., Research Fellow in Molecular Biology and Genetics, 1993-1999.
- University of California, Los Angeles, CA, U.S., PhD in Molecular Biology, 1993.
- University of British Columbia, BSc in Biochemistry, 1987.
MY RESEARCH OVERVIEW (GO TO SCIENTIFIC OVERVIEW)
Preserving the blueprint of life
How cells accurately transmit genetic information from one generation to the next is a fundamental biological problem and one that has held my interest since I was an undergraduate student.
In order to grow and divide, all living cells must duplicate their genetic material (DNA) and pass it accurately to the next generation. Failure to maintain a complete and error-free copy of the complete set of genes results in chromosome rearrangements, loss of genes, and accumulation of mutations—all hallmarks of cancerous cells and believed to be critically important in the development of cancers. My lab’s strategy is to use a simple organism to identify and study processes that may give rise to cancers in humans. We are studying two processes that are important for cancer development, progression, and treatment. The first is how cells copy their DNA without accumulating errors. The second is how cells deal with damage to their DNA, either from agents such as chemicals or radiation, or from processes within the cell itself, and how the cells are able to prevent that damage from leading to errors.
Understanding how DNA damage can arise and how cells respond to DNA damage is important for two reasons: DNA damage causes cancer, and DNA damage is used to treat cancer. New genes my lab identifies that cause increased DNA damage could be targets for future cancer treatments, or markers used to detect cancers. A better understanding of how cells respond to DNA damage is an important part of improving treatments, and in customizing them for individual patients.
The collaborative and multi-disciplinary environment of the Donnelly Centre encourages interactions with other scientists and the availability of cutting-edge technologies accelerates research. Science moves forward much more rapidly when researchers with complementary expertise contribute towards a common goal.
SCIENTIFIC RESEARCH OVERVIEW
Failure to maintain an error-free copy of the complete set of genes is a hallmark of cancerous cells, and is called genome instability. Accumulation of mutations, chromosome rearrangements, and loss of genes are all types of genome instability. Genome instability is central to understanding and treating cancer. The most common inherited forms of cancer (breast and colon) are caused by mutations in genes that prevent genome instability. Environmental agents that cause cancer often do so by causing genome instability. Genome instability is evident at the very earliest stages of cancer. Finally, the most common and successful anti-cancer treatments (aside from surgery) act by causing genome instability, either by damaging DNA or by interfering with its duplication. No area of modern biology is as intimately connected to understanding and treating cancer as is genome instability.
My lab’s strategy is to use a simple organism as a model system to identify and study processes that give rise to cancers in humans. Our model system offers robust genetics, a high degree of gene and pathway conservation with humans, ease of growth, and reduced complexity, all of which can rapidly expand our understanding of cancer gene function. In addition, our system offers a suite of cutting-edge tools that facilitate new approaches to understand biological complexity to an extent not yet possible in human cells. Our model system work is used as a springboard to understand the same processes and pathways in human cells.
Our studies contribute to understanding and treating cancer in three key ways. We plan to identify every gene that contributes to genome stability, ultimately providing a complete map of genes that can cause cancer, for use in diagnostics and personalized medicine. The new methods that we use to identify genome stability genes in our work will reveal conceptually novel targets for anti-cancer drugs. Our work will provide a better molecular understanding of how cells respond to genome instability, which is critical to improving existing therapies and customizing them for individual patients.
My lab has four main research nodes:
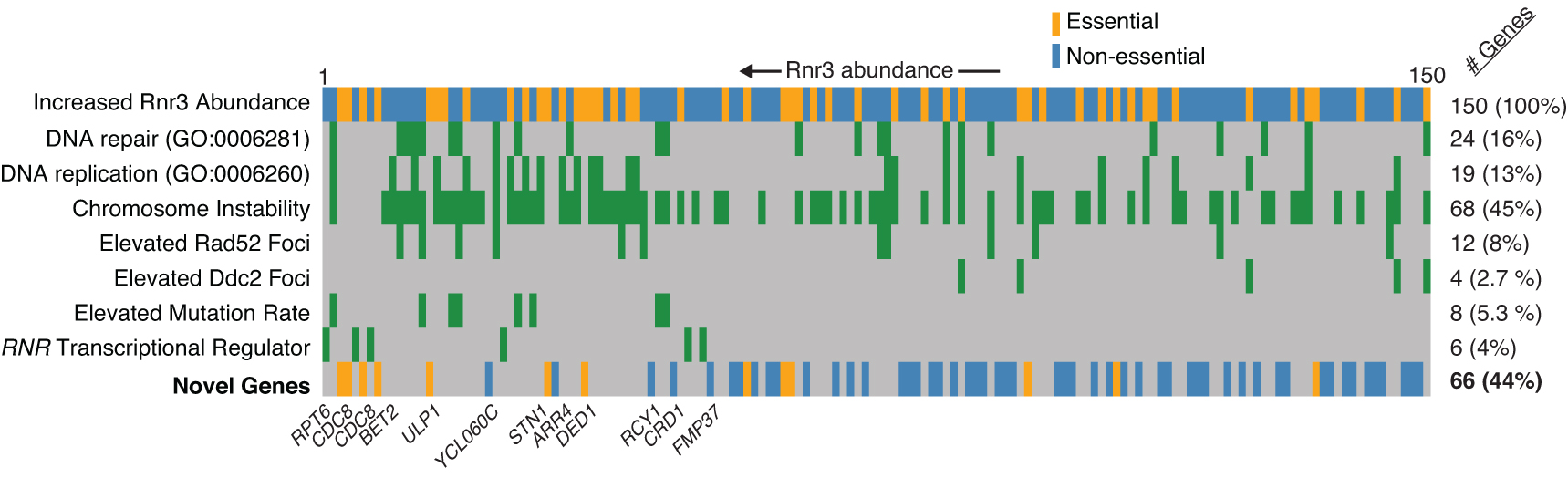
1. Genome instability gene and pathway identification by high-throughput, genome-wide, functional genomics screens in yeast
These studies use modern genetics and cell biology platforms to screen the yeast genome and proteome for genes and proteins that contribute to genome stability or that respond to genome damaging drugs. Current approaches include high content imaging screens in which hundreds of thousands of microscopic images are collected to track the location and abundance of every protein within individual living cells, and the use of a novel fluorescent reporter that identifies genome instability in individual cells.
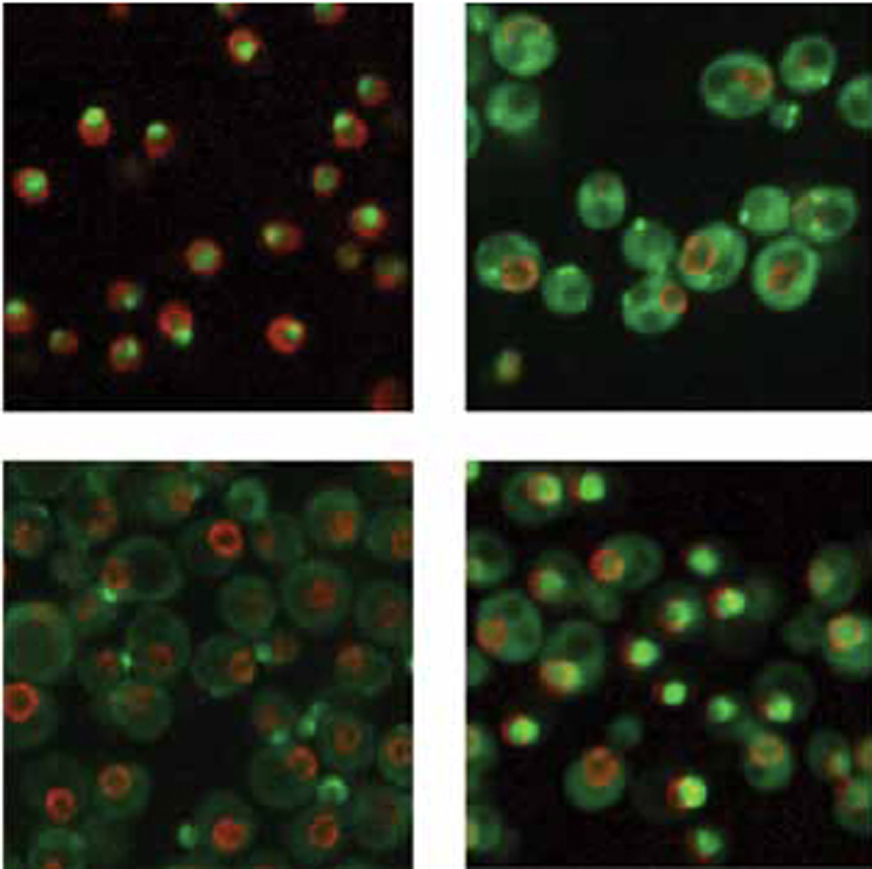
2. Analysis of protein re-patterning during DNA replication stress
 It has recently become clear that the most important causes of genome instability are barriers to genome duplication, referred to as DNA replication stress. We have used the intracellular movement of proteins during replication stress to identify new biological pathways that prevent genome instability during replication stress. We are now studying individual replication stress response proteins in both yeast and human cells, to understand how they function, and to learn if their function can be subverted as a cancer treatment modality.
It has recently become clear that the most important causes of genome instability are barriers to genome duplication, referred to as DNA replication stress. We have used the intracellular movement of proteins during replication stress to identify new biological pathways that prevent genome instability during replication stress. We are now studying individual replication stress response proteins in both yeast and human cells, to understand how they function, and to learn if their function can be subverted as a cancer treatment modality.
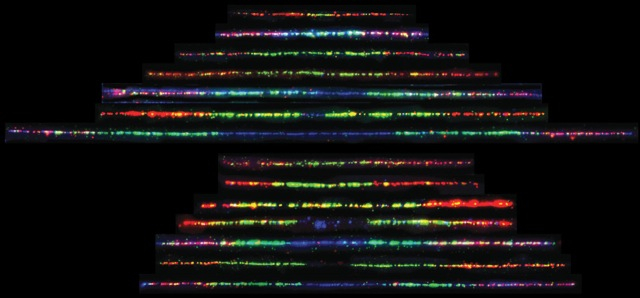
3. Understanding Bloom Syndrome
Bloom syndrome is a human cancer predisposition and aging syndrome that offers unique insight into the interplay between genome instability, DNA replication stress, and cancer. We are using modern proteomic techniques to identify human proteins that function in concert with the Bloom protein, and more traditional biochemical techniques to understand the function of a novel Bloom syndrome protein that my lab identified in a functional genomics screen several years ago.
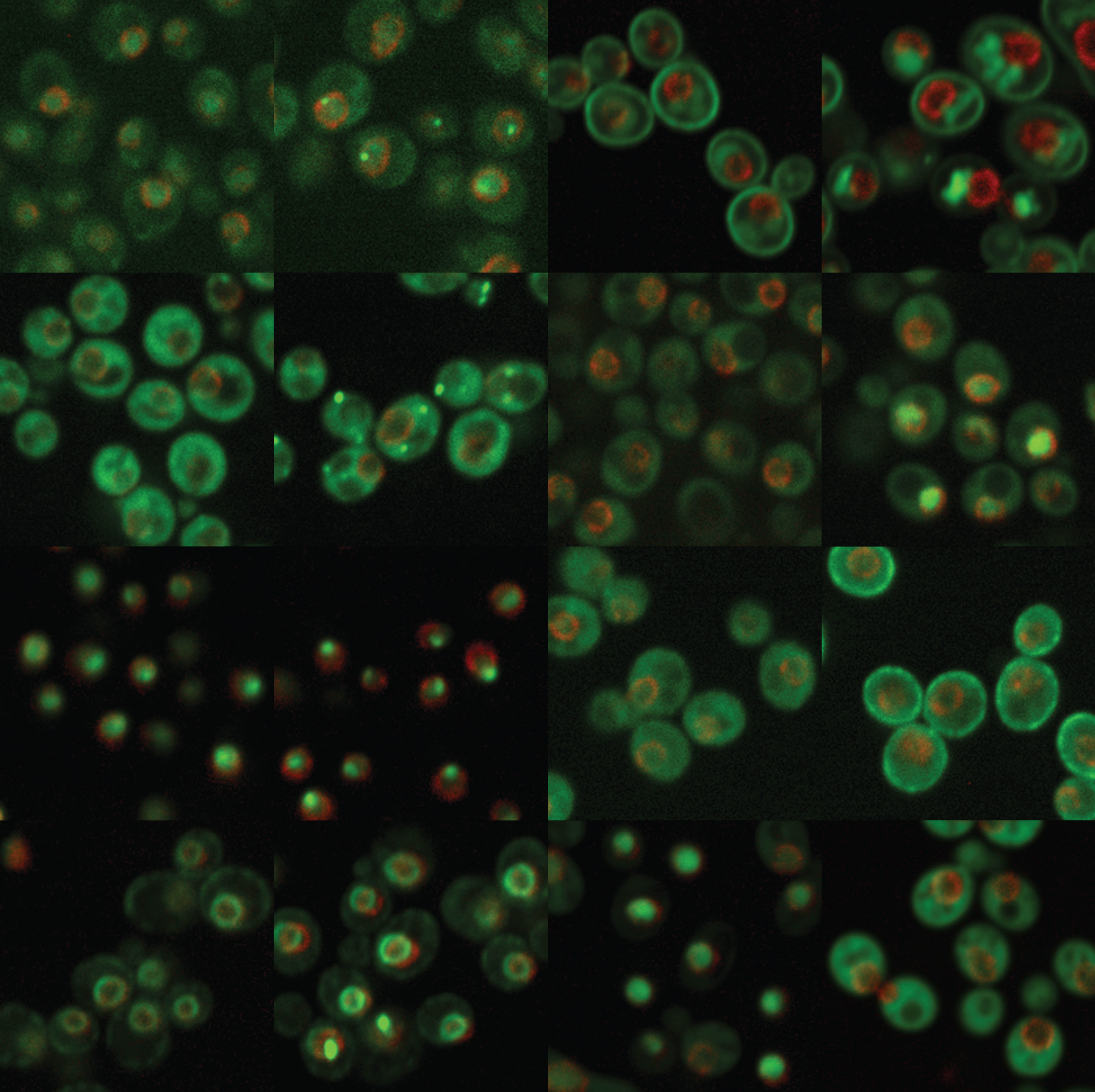
4. How cells maintain chromosome number
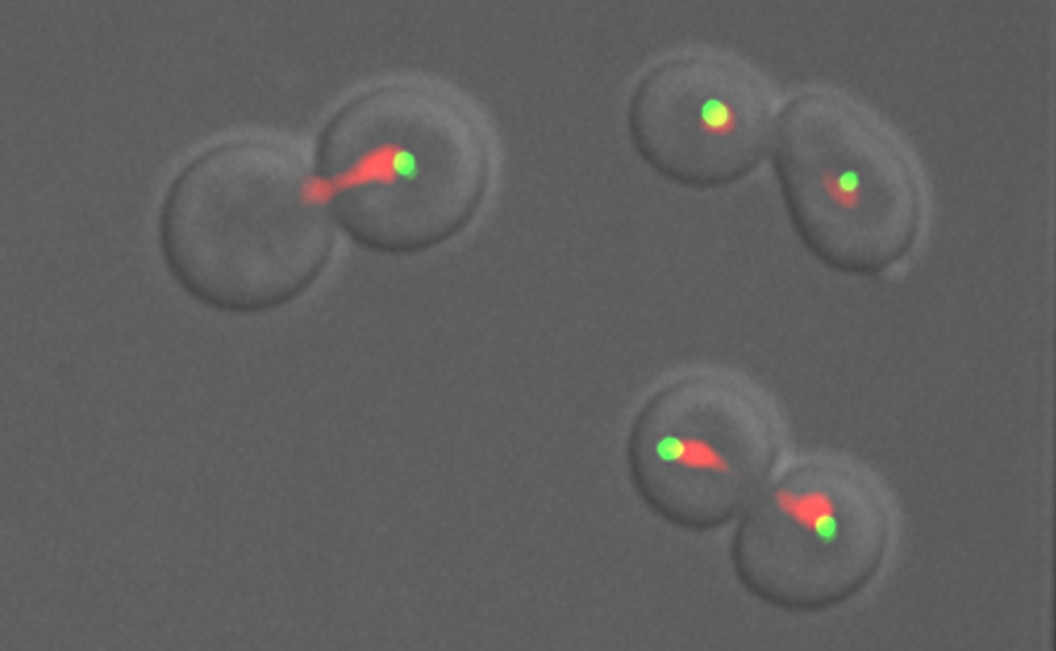 One hallmark of cancerous cells is changes in chromosome number. We have used flow cytometric and genetic techniques to identify all of the genes in yeast that are important for preventing increases in the number of chromosomes. We now seek a mechanistic understanding of how the genes and pathways we have identified operate.
One hallmark of cancerous cells is changes in chromosome number. We have used flow cytometric and genetic techniques to identify all of the genes in yeast that are important for preventing increases in the number of chromosomes. We now seek a mechanistic understanding of how the genes and pathways we have identified operate.
SELECT PUBLICATIONS
- Exploiting DNA Replication Stress for Cancer Treatment.Ubhi T, Brown GW. Cancer Res. 2019 Apr 15;79(8):1730-1739.
- Rad5 Recruits Error-Prone DNA Polymerases for Mutagenic Repair of ssDNA Gaps on Undamaged Templates.Gallo D, Kim T, Szakal B, Saayman X, Narula A, Park Y, Branzei D, Zhang Z, Brown GW. Mol Cell. 2019 Mar 7;73(5):900-914.
- Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome.Ho B, Baryshnikova A, Brown GW. Cell Syst. 2018 Feb 28;6(2):192-205.
